
The Biology Labs On-Line Project: Producing Educational Simulations That
Promote Active Learning
Jeffrey
Bell, California State University
Abstract
The Biology Labs On-Line Project is an attempt to create simulations of important
biology experiments that students normally cannot perform in typical undergraduate
laboratories. All of the simulations have enough complexity that students have
the flexibility to design and interpret their own experiments. The programs
generate large amounts of data and are fast enough for students to do multiple
experiments, creating the opportunity for students to get extensive practice
at applying scientific methods. The simulations are all written in Java and
are accessed over the World Wide Web, making them easily available to students
anytime and anywhere. Nine of the simulations, covering the topics of evolution,
Mendelian genetics, protein translation, human population demography, protein
structure-function, human genetics, mitochondrial electron transport, cardiovascular
physiology and photosynthesis are already finished, with six more planned for
the summer of 2000.
1. Introduction
1.1. Pedagogical
Motivation
A major lack in much current science
instruction at the university level is appropriate active learning experiences.
Students in many science courses still get too few opportunities to think and
reason about scientific problems. Well-designed laboratory experiments provide
the best means to give students the opportunity to learn about a science subject
while developing the thinking skills most instructors have as a goal of their
course. However, there are many limitations on the use of real laboratory experiments
in an undergraduate or high school science course. Students in educational labs
are severely limited by the time required for most serious investigations. A
typical laboratory for a biology class will meet once or twice a week for two
to three hours each time. This time constraint is a major barrier for introductory
students as they try to learn how to be a scientist. Many important biological
experiments take weeks, months or years to carry out, putting them well beyond
the reach of the typical teaching laboratory. Many other barriers limit the
choice of experiments in teaching laboratories, including a lack of appropriate
equipment, insufficient funds for expensive reagents, restrictions on the use
of hazardous chemicals and radioactive materials, a lack of technical skills
by the students and ethical concerns.
As a result of these difficulties, most professors either concentrate on using laboratory time to provide students with an opportunity to practice being a scientist by carrying out inquiry based investigations, or demonstrate scientific principles by having students carry out carefully controlled experiments designed by the instructor. In both cases the main mechanism for students to learn the subject is the traditional lecture. A major trend in current attempts to improve science education is to try to replace static lectures with more active learning approaches. While there are many versions of "active learning," in the sciences an inquiry approach is frequently used. One version of this is the learning cycle method in which students explore a biological phenomenon before receiving any explanation from the instructor. Other aspects of the inquiry approach include having the students propose their own hypothesis and design, execute and interpret their own experiments to test their hypothesis (Lawson, et. al, 1990, Uno, 1999). The National Research Council's Science Education Standards consider this the central strategy for teaching science (NRC, 1995). In addition to learning how to think like scientists, students learn the concepts and facts of a subject better when they have to apply the knowledge. While there are many ways to make the lecture more inquiry based, (see "Handbook On Teaching Undergraduate Courses" Uno, 1999), one very useful alternative is the use of simulations (Windschitl, 1998, Uno, 1999). To address these problems the Biology Labs On-Line Project has created several Java simulations of biological phenomena that can be used to supplement traditional laboratories and lectures, providing students with many more opportunities to learn by experimentation than is possible using traditional methods.
The Biology Labs On-Line Project is a component of the California State University System (CSU) Integrated Technology Strategy (ITS), which calls for anywhere, anytime access to information. The project initially brought together biologists from throughout the CSU system and the CSU Center for Distributed Learning (CDL) to explore ways to use technology to improve learning in introductory biology courses. Later, multimedia developers from Addison Wesley Longman were added to the development team. A major goal of the collaboration was to allow students to learn as biologists do, i.e., by actively designing experiments and interpreting their results. Eliminating the time constraints of the traditional experiment, the simulations give students the opportunity to design and interpret experiments, to learn from their mistakes, and to revise and redo their experiments just like real scientists. The simulations are not designed to replace the traditional "wet labs" found in the normal biology course, but rather to extend the laboratory experience to subjects and experiments that cannot normally be done, or not done well enough, in a traditional laboratory. The simulations are also not multi-media presentations, stand-alone tutorials or on-line courses.
A key advantage of a simulation is the potential of allowing the student to design and carry out many more experiments than would be possible with real labs. This gives the student many more opportunities to practice the skills of hypothesis creation, experimental design and data analysis than can happen in the normal lab or lecture setting. A lack of underlying complexity is a problem with many of the currently available online educational simulations, which frequently allow only one real experiment. Thus, one of the design goals for the Biology Labs On-Line Project was to create simulations with enough underlying complexity that students would be able to do many different experiments. For example, the FlyLab has 29 different genes on four different chromosomes, allowing the possibility of literally millions of experiments. Similarly, the Evolution Lab, which uses two islands, each with 7 independent continuously variable parameters, provides the capability of millions of different possible experiments. In addition to the complexity in the starting parameters, each of these programs operates stochastically, so that even with identical starting conditions students will get different results. The large number of possible experiments and the speed at which each experiment can be carried out, five to ten minutes per experiment, means that students can get much more practice at designing and interpreting their own experiments than is possible in the traditional laboratory. This creates new possibilities for teaching some topics as students can now figure out the underlying principle on their own, with only minimal guidance from their instructor.
The FlyLab’s introduction to the topic of sex linkage illustrates how these simulations can be used in inquiry. After doing some genetic crosses that demonstrate normal Mendelian dominance and recessive inheritance patterns, students are asked to investigate one of the X-linked traits available in the simulation. The students are not told that the trait is X-linked, nor is the concept of sex linkage discussed prior to the assignment. The students can do as many different crosses as necessary to try to figure out what is going on. The ability to carry out many different experiments is a key to making this work, as that is the only way the students can eliminate many of their initial explanations for what is happening. Even for the many students who fail to solve this problem, the experience is very helpful, making them much more attentive when the genetic explanation is given in class.
Another goal of the BLOL Project was to design simulations that could be used to learn about key concepts in biology that are not normally used in traditional laboratories because of time, expense, hazards, etc. Examples of this are the Evolution Lab and DemographyLab, which simulate processes that take place over hundreds of years; MitochondriaLab, which simulates experiments that use toxic chemicals; TranslationLab, which simulates the use of radioactive isotopes; HemoglobinLab and LeafLab, which simulate the use of complicated and expensive measuring equipment; PedigreeLab, which simulates expensive mapping experiments using dozens of human families; and CardioLab, which simulates potentially lethal experiments on human subjects.
1.2. Design of BLOL
and Comparison to Other Lab Simulations
Other laboratory simulation products exist,
comparable in some respects to the BLOL Project. Stella, for example, is a general
modeling tool, and Ecobreaker is a package that can be used to create useful
simulations of ecology processes. The Bioquest Consortium sells a large number
of simulations on CD, covering many of the same topics covered by BLOL. These
simulations differ from those in the BLOL Project, however, in that they have
certain machine requirements (Mac, PC, Unix) and distribution difficulties.
Our Java-based BLOL simulations, on the other hand, are platform independent
and not tied to use within the actual science laboratory.
The BLOL simulations have all been created in the Java programming language, so that they can be easily accessed over the web through any standard browser. This solves the problem of widely disseminating the applications, a common problem with most educational software. The Java application provides the user interface where students set the starting parameters for their experiment and get graphical feedback on their current settings. While each of the simulations is unique, all of them share many common interface elements and functions. In some of the simulations the Java program also calculates the results, while in others the input parameters are passed back to the server, where the real calculations take place. The students receive the results through the Java application.
The downside of using Java is that only individuals and schools with fairly new computers and software (Netscape 3 or better, etc.) and an internet connection can use the software. Another disadvantage of using Java is the inability of Java programs to save to disk or print. This limitation has been overcome through the use of a notebook that can be exported to a web page. All of the data tables, such as numbers of different types of progeny, or results of statistical calculations, can be imported directly into the notebook. After typing in their comments, students can export the notebook to a web page for printing, or to email to themselves or an instructor. The web page is temporarily stored on the server. Graphical images produced by some of the programs are also exportable to the notebook, where they can then be printed.
All of the programs share some common user interface elements, including a title bar with links to an introduction to the lab, help, sample assignments, the notebook, etc. While there is much diversity in how the different labs operate, most of them start in an input mode where the students design their experiment by adjusting different parameters, after which they run the simulation. The program calculates the results of the experiment, usually in a minute or less, and then presents the results in the output mode. In this mode there is a tabbed interface where the students choose which type of output they wish to view, a table of the data, a graph, the input values, etc. After analyzing their results they can import them into the notebook and then go back to the input mode to design another experiment. This ability to go back and forth quickly between the design of an experiment and the results is one of the powerful advantages of a simulation approach to teaching science.
1.3. Availability
For simulations to truly be useful as a primary
mechanism for teaching the concepts of a course, they need to cover the majority
of the key concepts in the course. The BLOL Project has produced nine different
educational simulations covering the subjects of evolution, Mendelian genetics,
protein translation, human population demography, protein structure-function,
human genetics, mitochondrial electron transport, plant photosynthesis and respiration,
and cardiovascular physiology. Current projects in progress and due to be finished
by the summer of 2000 will simulate enzyme kinetics (EnzymeLab), the use of
transgenic mice to study developmental genetics (TransgenicLab) , phylogenetic
reconstruction (CladisticsLab), population genetics (PopulationGeneticsLab),
metabolism (MetabolismLab) and population ecology (PopulationEcologyLab). Thus,
a significant portion of the major concepts introduced in a typical introductory
biology course could be taught using a combination of real laboratories and
these simulations.
Below is a brief description of the finished simulations.
2. DemographyLab
The Demography Lab models human population
growth for seven different countries around the world. Students can use this
lab to investigate how differences in population size, age-structure, and age-specific
fertility and mortality rates affect human population growth. Default values
for seven countries-- China, India, Japan, Mexico, Nigeria, Sweden, and the
United States-- have been incorporated into the program to allow comparisons
between nations with very different demographics, such as Japan and Nigeria.
In addition, students can change any of the parameters to create their own experiments.
Initial population size can be set anywhere from two to six million. The proportion
of males and females in each five year age group can be set independently of
one another. Thus, the effects of a war or a disease that affects one sex or
certain age groups can be modeled. The mortality rate for males and females
in each five year age group can also be set independently so the effects of
changes in health care can be modeled (by changing infant mortality rates or
mortality rates for the elderly, for instance). The birth rate per female in
each five-year age group can all be set for those ages in which birth rates
are possible. Thus, the effects of birth control efforts or changes in marriage
ages can be modeled. All of these parameters are adjusted using a simple graphical
interface.
After running the simulation for 100, 200 or 300 years, students get summary statistics, such as life expectancy, birth rate, population growth rate, etc. They can view a line graph of population numbers over the course of their experiment, see a graphical representation of the population structure for every five years of the experiment, or examine the number of males and females in each age group for each five year period. With the program a variety of demographic phenomena can be demonstrated, such as exponential growth and decline, stable age structure, zero population growth, demographic momentum, dependency ratios, sex ratios and marriage squeezes.
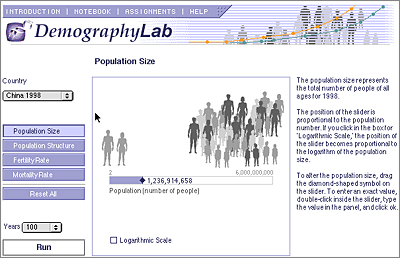
Figure 1. A thumbnail of the DemographyLab screenshot.
![]() A QuickTime movie (533 KB) showing how the DemographyLab works.
A QuickTime movie (533 KB) showing how the DemographyLab works.
![]() A screenshot (16 KB) showing the DemographyLab.
A screenshot (16 KB) showing the DemographyLab.
3. EvolutionLab
Although evolution is the unifying theme
of the biological sciences, it is perhaps one of the most misunderstood and
difficult concepts to convey in a laboratory setting. The study of evolution
is especially suited to computer simulations because evolution normally occurs
over very long time intervals, large data sets are usually needed to understand
it, and there are usually a number of important parameters that are difficult
to control in real experiments. EvolutionLab is a web based, interactive computer
simulation designed to teach the basic concepts of natural selection and to
convey the importance of time in the evolutionary process.
Students using EvolutionLab observe evolutionary changes in bird beak morphology in hypothetical populations of birds isolated on two islands. In the simulation, students can set the annual rainfall on island(s) containing finch populations, and then observe the effect of this environment on the evolution of the finches’ beaks. Students may also change several other properties of the bird populations, such as initial mean beak size, beak size variability, beak size heritability and mean clutch size, to determine their effect on beak evolution.
While the simulation is based on Darwin’s finches, changes in the species variables such as mean beak size, variability, heritability and clutch size create virtual species that can have properties similar to many other wild species. Students can investigate the parameters that are more likely to lead to the extinction of endangered species, see why some species might evolve faster than others, and examine many other facets of evolution. The program generates large data sets -- one run can produce 600 data points -- so students can learn how to analyze and interpret large amounts of data, unlike the situation in a typical lab. The great flexibility of the program should allow individual instructors to tailor student assignments to their particular preferences and provide students with a real opportunity to design their own experiments. Actively engaging students in exploring and studying evolution through this simulation provides another avenue for students to learn about evolution in addition to the traditional text and lecture explanations.
![]() A QuickTime movie (561 KB) showing how the EvolutionLab works.
A QuickTime movie (561 KB) showing how the EvolutionLab works.
![]() A screenshot (18 KB) showing the EvolutionLab.
A screenshot (18 KB) showing the EvolutionLab.
![]() A QuickTime movie (663 KB) showing how the PedigreeLab works.
A QuickTime movie (663 KB) showing how the PedigreeLab works.
![]() A screenshot (23 KB) showing the PedigreeLab.
A screenshot (23 KB) showing the PedigreeLab.
5. FlyLab
The FlyLab simulation is an update to
the FlyLab originally created by Bob Desharnais. In the FlyLab, students design
their own fruit flies by choosing from many different possible phenotypes for
characteristics such as eye color, wing shape, body color, etc. They then mate
their flies and analyze the progeny to determine the rules of inheritance for
different traits. Each experiment is unique, and students can have up to 10,000
progeny produced from one mating. Offspring can also be mated, providing a wide
range of different experiments. There are 29 different traits that can be studied
in isolation or in various combinations so the number of possible experiments
is in the millions. The traits are all represented graphically, allowing the
student to observe the phenotypes directly. For instance, if the student selects
the white eye mutation for the female parent, the picture of the female parent
will have white eyes. After the mating, pictures of the different progeny are
presented, along with numbers beside each picture to indicate the number of
progeny of that type (number of females with white eyes, females with red eyes,
etc.) The program includes a Chi Square calculator for doing statistical tests
of the students’ hypotheses, and a notebook for recording results, observations,
hypotheses and conclusions. Students can import the numerical results from their
crosses and statistical tests directly into the notebook.
Using this program, students can discover or study most of the important principles of Mendelian genetics, including dominant and recessive alleles, sex-linkage, lethal alleles, independent assortment, epistasis, linkage, gene order, linkage groups, and linkage maps. An advantage of the simulation is that students can discover these principles by doing the same sort of experiments as the original researchers, only much more quickly. The program is appropriate for a wide range of biology courses as the assignment determines the level of difficulty. Students can do statistical tests, but this is not required. They can do complicated crosses with multiple traits, or simple crosses with only one trait at a time. If students are confused by a complicated cross, they can always do some additional simpler crosses to try to figure out what is going on. They can also do additional crosses with the progeny from their crosses, and their progeny, etc. This ability to devise their own experiments and try many different permutations is a major strength of the FlyLab.
![]() A QuickTime movie (1.46 MB) showing how the FlyLab works.
A QuickTime movie (1.46 MB) showing how the FlyLab works.
![]() A screenshot (23KB) showing the FlyLab.
A screenshot (23KB) showing the FlyLab.
6. TranslationLab
So far, the BLOL project has produced two
molecular biology simulations. The first, TranslationLab simulates some of the
original experiments used to crack the genetic code, one of the key discoveries
in molecular biology. These experiments rely on radioactive materials and difficult-to-produce
RNA templates, so they can't be done in the normal biology lab. Students design
and create simple RNA molecules in the simulation that they then translate in
a virtual in vitro translation mix. The program shows a simple animation
of the techniques that would be used to analyze the products of the translation
and then gives them the amino acid sequence of any proteins produced in their
experiment. The student must logically analyze the results of multiple experiments
to deduce the properties of the genetic code, just as the original researchers
did, only with the advantage of being able to do experiments in minutes that
normally take months to carry out. Various properties of the code that can be
determined using this simulation are the triplet nature of the code, the non-overlapping
nature of the code, the degenerate nature of the code, codon assignments for
each amino acid, and the existence and identity of stop codons.
![]() A QuickTime movie (1.45 MB) showing how the TranslationLab works.
A QuickTime movie (1.45 MB) showing how the TranslationLab works.
![]() A screenshot (39 KB) showing the TranslationLab.gif.
A screenshot (39 KB) showing the TranslationLab.gif.
![]() A QuickTime movie (751 KB) showing how the HemoglobinLab works.
A QuickTime movie (751 KB) showing how the HemoglobinLab works.
![]() A screenshot (21 KB) showing the HemoglobinLab.
A screenshot (21 KB) showing the HemoglobinLab.
![]() A QuickTime movie (2.08 MB) showing how the MitochondrialLab works.
A QuickTime movie (2.08 MB) showing how the MitochondrialLab works.
![]() A screenshot (38 KB) showing the MitochondriaLab.
A screenshot (38 KB) showing the MitochondriaLab.
![]() A QuickTime movie showing how the LeafLab (1.61 MB) works.
A QuickTime movie showing how the LeafLab (1.61 MB) works.
![]() A screenshot (24 KB) showing the LeafLab.
A screenshot (24 KB) showing the LeafLab.
![]() A QuickTime movie (946 KB) showing how the CardioLab works.
A QuickTime movie (946 KB) showing how the CardioLab works.
![]() A screenshot (24 KB) showing the CardioLab.
A screenshot (24 KB) showing the CardioLab.
The precursor to all of these labs is the original Virtual Fly Lab. This simulation of fruit fly genetics is now used in biology classes all over the world, and has created so much demand on the server hosting the program that there are now five different mirror servers. I field tested the FlyLab, EvolutionLab and TranslationLab in an upper division genetics course with encouraging results. 98% of the students in this course considered their FlyLab assignments useful in learning genetics; 83% found EvolutionLab to be useful; and 93% found TranslationLab useful. Some comments from the students are given below (Student comments refer to older names for the applications. Virtual Fly is now FlyLab, EvolveIt is now EvolutionLab and TranslateIt is now TranslationLab).
"Virtual Fly and TranslateIT were the assignments I got the most out of. I liked the way it made you systematically think to solve the problems."
"TranslateIt was enjoyable because it requires the student to investigate and solve the problem."
"TranslateIT gave me an excellent concept of how genes code for proteins through valuable experience. I could experiment and learn by trial and error to prove to myself that the book and what I was working on were in fact the same."
"It is a tie between TranslateIT and EvolveIT for most useful and as to how much I got out of them. They were easy to use, the help info was well defined and they both helped me see the larger concepts, 'big picture'."
"I liked the Virtual fly and EvolveIt activities because they allowed you to do some investigation on your own and they made you think about what was really happening, which made you understand the material better."
There were only a few negative comments, usually having to do with the difficulty of getting on-line and using the programs. Students who are uncomfortable with computers are at a disadvantage when using these simulations and special care must be taken to make sure they get the most out of the simulations. The only other negative comment was, "I liked TranslateIt the least because it made my head hurt." While this is unfortunate, if the BioLabs project can produce more simulations that cause some students’ heads to hurt, then the project will be producing simulations that change, for the better, the way biology is taught.
12. ConclusionJensen, M. S. 1998. Finding a Place for the Computer in the Introductory Biology Laboratory. Journal of College Science Teaching 38(2): 247-249
Lawson, A. E., Rissing, S. W., and Faeth, S. H. 1990. An Inquiry approach to Nonmajors Biology. Journal of College Science Teaching 30(5): 340-346
National Research Council. 1990. National Science Education Standards. National Academy Press, Washington, D. C.
Uno, G.E. 1999. Handbook On Teaching Undergraduate Courses. Saunders College Publishing, Orlando, Florida
Windschitl, M. A. 1998. A Practical Guide for Incorporating Computer-Based simulations into Science Instruction. American Biology Teacher 60(2): 92-97
14. Acknowledgements
The following individuals contributed to the development
of these simulations. Their contributions are gratefully acknowledged: Bob Desharnais,
David Caprette, Mike Palladino, Steve Wolf, Zed Mason, Ron Quinn, Terry Frey,
David Hanes, Judith Kandel, Nancy Smith, Sally Veregge, Abbe Barker, Michelle
LaMar, Mark Crowley, Chuck Schneebeck, Rachel Smith, David Risner, Swann Do,
Lou Zweier, Scott Anderson, Peilin Nee and Anne Scanlan-Rohrer. Partial support
was provided by U.S. National Science Foundation grant DUE 9455428 to Bob Desharnais.
********** End of Document **********
IMEJ multimedia team member assigned to this paper: Yue-Ling Wong